How to analyse P2
Protocol 2
The data from protocol 2 is processed using the function process_protocol2(). This function should be called within the experiment folder that was created when generate_protocol2() was run. It should follow the format data/YYYY_MM_DD_HH_MM.
This function reads in the metadata file that has information about the strain of the fly, peak_frame, side of the arena.
The responses to the moving bar stimuli are processed first, since this analysis generates a vector sum of the responses of the cell to bars moving in the 16 different directions.
The resultant angle of this vector sum is then fed in to the function that processes the flash responses and overlays an arrow with this direction on top of the receptive field plots.
Bar sweep analysis: Produces polar plots of the responses to bars moving in 16 different directions, as well as a vector sum arrow that indicates the preferred direction of motion of the cell.
Flash analysis: Produces heatmaps of the responses to the 4 pixel square flashes at both 160ms and 80ms durations, for both ON and OFF contrasts. An arrow is overlaid on these plots that indicates the preferred direction of motion found in the bar sweep analysis.
How the data is organised
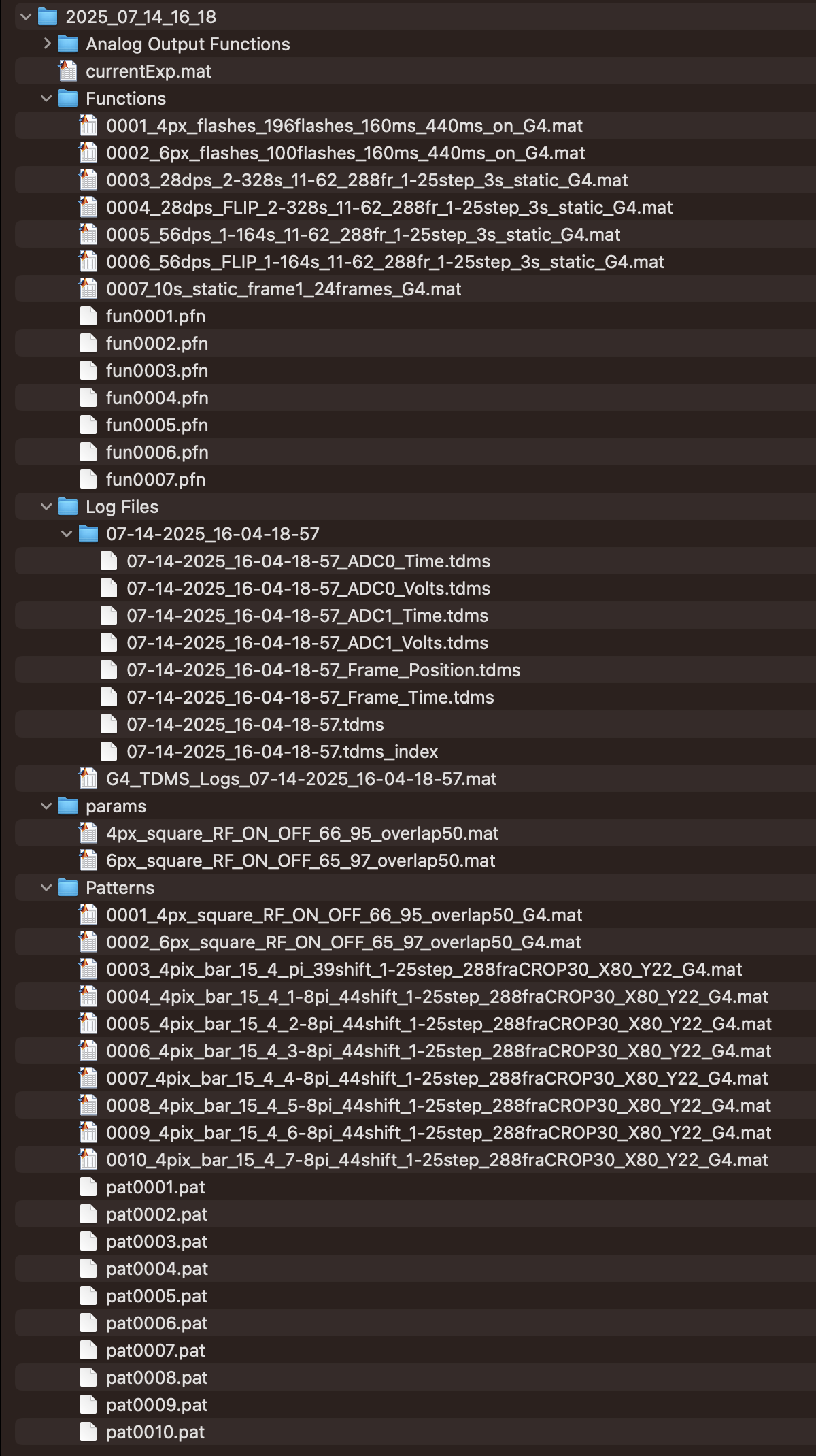
- Each run of
generate_protocol2()with create a new folder withinC:\matlabroot\G4_Protocols\nested_RF_protocol2with the format “YYYY_MM_DD_HH_MM” of the date and time when the protocol was run. - Within this folder there will be a
patternsfolder with the patterns that were used in the protocol, afunctionsfolder with the position functions that were used, and aparamsfolder containing a file with metadata about the protocol run. - There will also be a folder
Log Files, this itself will contain a subfolder with the exact date and time of the run that will contain the raw tdms files of the electrophysiology data and the frame position data. Witingenerate_protocol2()there is a functionrun_protocol2()that runs the protocol created. At the very end of this function there is a function calledG4_TDMS_folder2struct(log_folder)- another function from theG4_Display_Toolsthat will convert the tdms files into a combined structure (calledLog) that can be used for analysis.