Flash analysis
Breakdown of process_flash_p2
After the responses to the bar stimuli have been processed the function src/analysis/protocol2/process_flash_p2 is used to process the responses to the two different flash stimuli. This function takes in the parameter resultant_angle from process_bars_p2. At the beginning of this function, the frame position data and the voltage data are once again loaded in. The median voltage across the entire experiment is calculated and the voltage data is also downsampled with movmean(v_data, 20000). The function then runs through the data for the two different sizes of flash stimuli separately.
Parsing the flash data
First, the function src/analysis/protocol2/pipeline/parse_flash_data is used to parse the frame data to understand when the flash stimuli were presented. This functions requires on_off, slow_fast and px_size as input parameters to know whether bright ‘on’ or dark ‘off’ stimuli were presented, whether the ‘slow’ or ‘fast’ flash speed was used and the size of the flash in pixels, respectively. This function returns several square arrays all of size [n_rows, n_cols] that contain individual values per flash position.
cmap_id- Each flash position is assigned the value either 1,2 or 3. 1 = larger excitatory peak, 2 = larger inhibitory peak, 3 = no peak big enough to be classified as 1 or 2.data_comb- Depending on the value ofcmap_id(r, c)this will either be the maximum voltage (cmap_id = 1), minimum voltage (cmap_id = 2) or the mean voltage across the last 25% of the flash. This value is used to set the intensity of the colour for that flash position in the heatmap plot. See the functionplot_rf_estimate_timeseries_lineandplot_heatmap_flash_responses.var_across_reps- Mean coefficient of variance across repetitions. How reliable is the response?var_within_reps- Mean coefficient of variance within repetitions. How different is the response from baseline?diff_mean- Maximum voltage during flash response - minimum voltage during flash response.max_data- 98th percentile value during flash presentation (TODO)min_data- 2nd percentile value during second half of flash presentation (TODO - include interval time after too).
- Find the range of timepoints during which all of the flash stimuli for each repetition were being presented.
Similar to how the bar data is parsed, the difference between the frame positions is used to coarsely divide the data up into the different stimuli. For the flash stimuli the variable idx contains the timepoints that correspond to the start of the different flash stimuli repetitions (both the 4 pixel and the 6 pixel flashes).
As a reminder, P2 consists of 4 pixel flash stimuli presented, followed by 6 pixel flash stimuli of one contrast (designated by the value of the parameter on_off). Both the ‘on’ and the ‘off’ versions of P2 use the same pattern files that contain frames with dark flashes followed by frames with bright flashes. It is only the position functions, the files that tell the controller which frame of the pattern to present, that differ between the ‘on’ and ‘off’ versions of P2. Because of this, a key difference then between the ‘on’ and ‘off’ versions of P2 is that the first flash of both the 4 pixel and 6 pixel flashes for the ‘off’ version is frame 2 of the pattern - which is equal to the value 1 in f_data. Whereas in the ‘on’ version of P2, the first flash of the 4 pixel flashes is f_data = 197 and the first flash of the 6 pixel flashes is f_data = 101. This is because there are 196 4 pixel dark flashes and 100 6 pixel dark flashes in the pattern before the bright flashes. This information is used to find the start of the 4 pixel flashes and the 6 pixel flashes.
To find the end of the flash stimuli repetitions, the beginning of the 6 pixel flash stimuli is used to set the end of the 4 pixel flash stimuli. The end of the 6 pixel flash stimuli is found using the same method that is used when parsing the bar data. These timepoint indices are stored in idx_6px.
[ PNG - 0014 ] ^ f_data with vertical lines for the beginning of each flash stimulus - OFF STIMULUS.
[ PNG - 0015 ] ^ close up of 0014 for the start of 4px flashes - rep 1.
[ PNG - 0015-5 ] ^ f_data with vertical lines for the beginning of each flash stimulus - ON STIMULUS.
- Find the timepoints for when individual flash stimuli start and stop.
start_flash_idxs is then generated that contains the timepoints within the range of the flash repetitions where the difference in frame position is >0 - this happens whenever a flash starts. These indices are then used to extract the frame data and voltage data per flash. For each flash, the data extracted includes 1000 datapoints before the flash starts until 6000 datapoints after the flash starts. As of July 2025, both the 4 and 6 pixel flashes are presented for 160ms then have a 440ms interval before the next flash starts. 160+440 = 600ms = ~6000 datapoints. So, the data extracted includes 1000 datapoints before the flash starts until just before the next flash starts. The timeseries of frame data is stored within the array data_frame of size [n_reps, 7000], the median-subtracted voltage data is stored in the array data_flash of size [n_reps, 7000] and the raw voltage data is stored in the array data_flash_raw of size [n_reps, 7000].
[ PNG - 0016 ] ^ Red line shows the start of the first flash Cyan line shows the actual range of timepoints for the first flash - 1000 timepoints before to 6000 timepoints after.
- Compute metrics across flash reps and for the mean flash response.
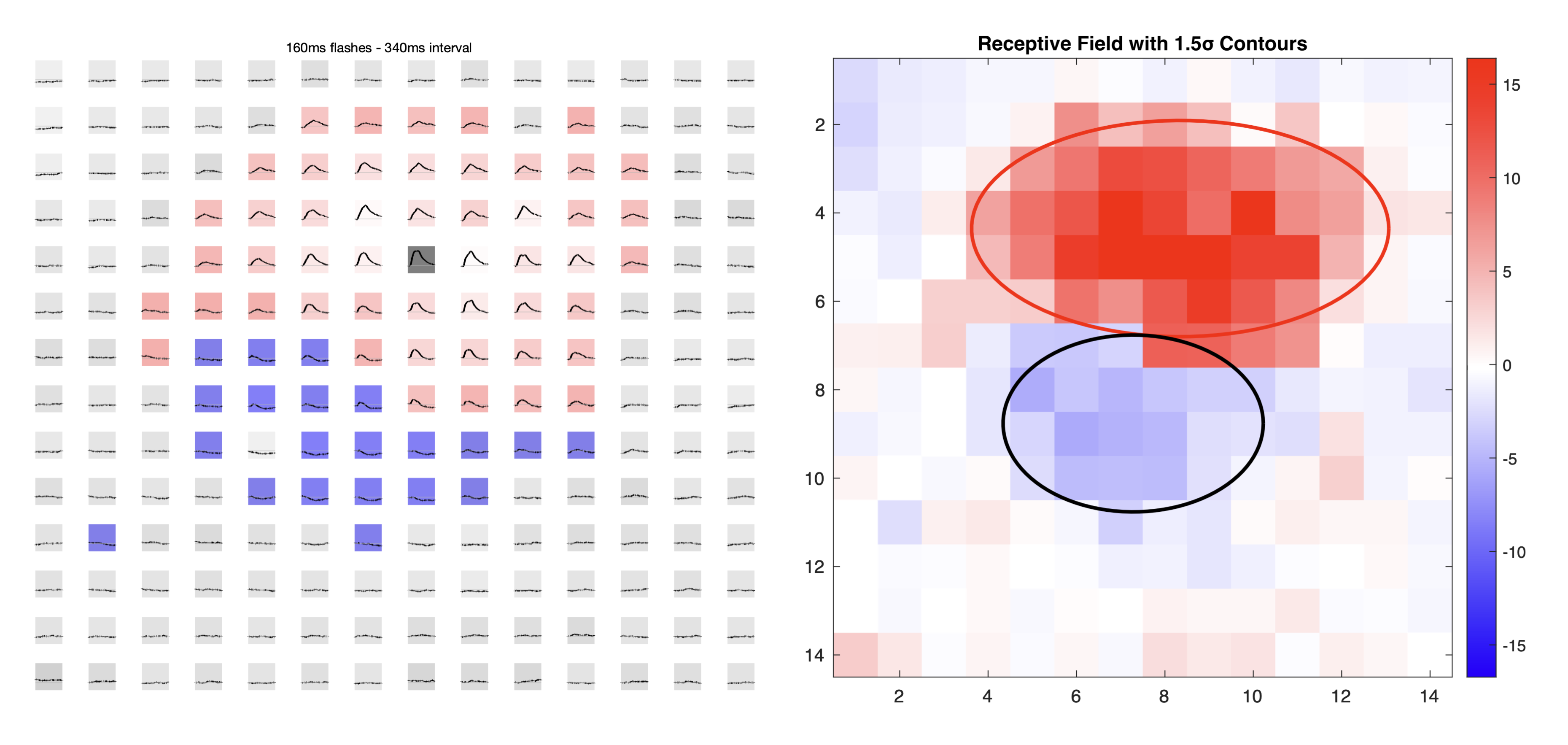
A number of metrics are then calculated for each individual flash position including the variance across and within reps, the maximum and minimum voltage during the rep and the difference between the maximum and the minimum response. These metrics are then used to assign each flash position to a cmap group for later plotting. Basically, flash positions that showed a large depolarising response get assigned to group 1 and will have a warm colourmap in the heatmap plot, flash positions in group 2 have a large hyperpolarising response and will have a cool colourmap and flash positions that don’t have a big enough difference in their maximum and minimum voltage across the flash presentation are assigned to group 3 and will be white in the heatmap.
Plotting the flash data
Within process_flash_p2, the code loops through the 4 pixel size flashes and then the 6 pixel size flashes. It makes two receptive field plots for both and stores metrics about the voltage data and the receptive field size and shape in a nested structure rf_results.
- Grid format plot with the mean timeseries of each flash response and the subplot’s background colour-coded by the
cmap_idvalue anddata_combvalue. (src/analysis/plotting/plot_rf_estimate_timeseries_line)
This figure contains the mean voltage timeseries per flash position organised by its location in space (corresponding to its position on the screen from the fly’s perspective). The voltage data is downsampled by 10 to try and reduce the size of the figure slightly. If a flash position subplot is assigned cmap_id of 1 or 2 then it’s background is filled with a uniform red or blue background, respectively. The intensity of the background is proportional to that flash position’s data_comb value that is linked to it’s absolute maximum or minimum voltage. These values were normalised between 0 and 1 before passing the array into the function. So, flash positions with the highest maximum voltages will have the darkest red backgrounds and flash positions that elicited the lowest minimum voltage will have the darkest blue backgrounds. Flash positions with cmap_id value = 3 will have white / grey backgrounds. The resultant_angle from the processing of the bar stimuli is passed through to this function and an arrow is overlaid on top of the subplots, positioned within the centre of the figure and pointing in the direction of the resultant_angle.
[ PNG - 0017 ] ^ Timeseries heatmap plot
- Heatmap plot (
src/analysis/plotting/plot_heatmap_flash_responses)
This function creates a heatmap of the normalised values within data_comb (values between 0 and 1) using a redblue colourmap. The colourmap is centred around the median value to get a good colour range and so that the median value is white.
med_val = median(data_comb(:));
clim([med_val-0.5 med_val+0.5])[ PNG - 0018 ] ^ Heatmap plot
Calculating metrics from the flash data
The function src/analysis/protocol2/gaussian_RF_estimate fits and plots a rotated 2D Gaussian fit around the excitatory and inhibitory lobes of the receptive field. It takes in the normalised and unnormalised versions of data_comb as it’s inputs and uses the built in MATLAB function lsqcurvefit to optimise the parameters for the gaussian fit. The function generates multiple plots showing the fitted data for both the excitatory and the inhibitory lobes and returns the metrics:
optEx-R_squaredoptInhR_squaredif1- fitted inhib plotf2- fitted excite plot
[ PNG - 0019 ] ^ fitted excitatory plot
[ PNG - 0020 ] ^ fitted inhibitory plot